General Research Topics
- Development of databases and database integration
- Information systems for metabolic diseases
- Biological paradigms of computability
- Conceptual datastructures for semantic database integration
- Modeling and simulation of metabolic processes
- Parallel computing for sequence analysis
- Multimedia implementation of virtual scenarios
- Bio-Medical knowledge processing
Research Projects
| BMBF Project: A Datawarehouse for the reconstruction and analysis of Micro-RNA metabolic network control |
|
Bioinformatics modelling and simulation of biological networks are one of the most important areas in system biology. Biological networks are powerful frameworks to integrate experimental as well as database knowledge in order to model biological interactions and systems. Therefore, different biological networks exist, such as protein-protein-interaction networks, signalling networks, gene-regulation-networks, and metabolic networks, among others. However, the fine- and fast-response regulation mechanisms within cells, like realized by microRNAs, have not been modelled, either simulated due to the lack of biological data. MicroRNAs are one of the most important mechanisms in the fine-regulation of meany biological processes. Many reports have already shown, that this new –omic level opens the door to a completely new cell mechanisms. Although, the investigation on microRNAS is still in its beginning, it was possible to demonstrate, how big the impact of microRNA regulation can be. Right now, data about microRNA is very limited and not connected to other –omic level showing its regulation mechanisms. Therfore, we aim to build a new microRNA database, which can be linked to other life-schience databases in a datawarehouse approach. The aim is to reconstruct microRNA-metabolic networks showing the impact of various microRNA, whoich later on can be simulated. Therefore, our project partner in Izmir will revise their existing RNA data and made this information accessible in a new relational database. This database will be the base for the modelling, analysis, and reconstruction of the biological networks in Bielefeld. Computer scientists from Bielefeld will integrate the new microRNA database into an existing database called DAWIS-M.D. where it will be semantically connected to other life-science databases, such as KEGG, HPRD, MINT, and IntAct, among others. This will result in biological correlation covering the most important –omic level. Once integrated in the datawarehouse, the modelling tool VANESA will be extended in its functionality to reconstruct, analyse, and automatically simulated the microRNA-metabolic networks in the Petri net language. Thus, using VANESA scientists will have the possibility to predict complex networks from RNA particles, extend existing networks with RNA fragments, and/or extend already known networks from literature with RNA information. Besides the technical realization of this novel approach, we aim to investigate the microRNA-metabolic networks in the context of our cardiovascular networks, which were reconstructed without microRNA interference in previouse work. Up to now, it was not possible to integrate this new level of micro-RNA fine regulation. Another goal we aim to realize is the tissue-specific reconstruction of those networks. With additional data from our partners from Izmir it is planned to reconstruct a 3D cell in the software application CellMicrocosmos, showing the regulation range of microRNA. Especially the membrane and nucleous transport is of great interest. With the data from the datawarehouse we are already able to determine the location of many different genes. In combination with the microRNA we will try to show the specific location of regulation and moreover the time of regulation. This will lead to new insights in fundamental research. |
| BMBF/ESF Project: KALIS - Ein webbasiertes Informationssystem zur patientenindividuellen Arzneimittel-Risikoprüfung |
|
Das medizinische Personal und die Patienten haben im Zeitalter der Mehrfachmedikation die Herausforderung Wechsel- und Nebenwirkungen von Arzneimitteln zu minimieren, da diese sowohl in hohen und unvorhersehbaren Kosten, als auch in unerwünschten teils lebensbedrohlichen Konsequenzen resultieren. In dem geplanten Vorhaben wollen wir angelehnt an diese Problematik ein webbasiertes Informationssystem zur Identifikation von Arzneimittelwechselwirkungen und neuen unerwünschten Arzneimittelwirkungen realisieren. Dieses System weist nicht nur auf mögliche Risiken mit bereits bestehender Medikation hin, sondern auch auf mögliche Wechselwirkungen zwischen Medikamenten und Lebensmitteln. Zusätzlich ist es in der Lage, eine sinnvolle Alternativtherapie und homöopathische Mittel zu bestimmen. Außerdem ermöglicht ein integrierter Algorithmus eine beobachtete Nebenwirkung einem oder mehreren Bestandteilen der Medikation zuzuordnen. Auf diese Weise wird das Risiko in der Arzneimitteltherapie gemindert und eine Kosten- und Zeitersparnis ermöglicht. Zudem gewährt das System einen Einblick in die darunterliegenden biomedizinischen Netzwerke, die für die Wissensgewinnung und Aufklärung eine fundamentale Rolle spielen. Denn eine Krankheit wird nicht nur durch ein Bakterium, ein Virus oder auch ein Gen hervorgerufen. Vielmehr handelt es sich hier um das Zusammenspiel vieler komplexer biochemischer Netzwerke. Run time: 2013 - 2014 Project Members:AG Bioinformatik
Prof. Ralf Hofestädt |
| MoRitS: Modellbasierte Realisierung intelligenter Systeme in der Nano- und Bio-Technologie |
|
|
| BMBF Project: Development of algorithms for bioinformatics analysis of complex metabolic and molecular-genetics networks |
German Partners:Prof. Dr. Ralf Hofestädt, Bielefeld University, Bioinformatics/Medical Informatics Department Prof. Dr. Falk Schreiber, IPK Gatersleben, Plant Bioinformatics Group Russian Partners:Prof. Dr. Nikolay Kolchanov, Institute of Cytology & Genetics, Siberian Branch of the Russian Academy of Sciences, Novosibirsk, Russia Dr. Ivanisenko, PBSoft Ltd., Novosibirsk Russia |
| BMBF Project: Deutsch/Russisches Forschungs-Netzwerk: Integrative Analyse komplexer metabolischer Netzwerke |
German Partners:Prof. Dr. Ralf Hofestädt, Bielefeld University, Bioinformatics/Medical Informatics Department Russian Partners:Prof. Dr. Nikolay Kolchanov, Institute of Cytology & Genetics, Siberian Branch of the Russian Academy of Sciences, Novosibirsk, Russia |
| DAAD Project: Leonhard-Euler-Programm: Integrative Bioinformatics |
|
Name: Bragin Anatoly Olegovich
Name: Tiys Evgeny Sergeevich
Name: Ivanisenko Timofey Vladimirovich
|
| BMBF Project: "Semi-automatic construction and visualization of complex metabolic networks based on the Petri net Concept" |
|
Scientific objectives The main goal of Systems Biology world-wide is to realize the implementation of a virtual cell. However, the electronic infrastructure for the realization of this fundamental idea does not yet exist.The main goal of this cooperative project is to develop and implement a tool for the semi-automatic implementation of complex metabolic Petri nets. Therefore, we have to implement a data integration tool, which will allow the automatic access to relevant metabolic databases (KEGG, BRENDA, TRANSFAC, TRANSPATH, BIND, etc.). Based on this integration layer, we will implement a visualization tool for complex metabolic networks and a Petri net transformation tool, which is able to translate selected metabolic networks into the language of Petri nets. In case of the Petri net simulation, the outstanding Petri net simulation tool Cell Illustrator will be integrated. Based on this tool, we will test and evaluate the system using data from rice (coming from the Chinese partner) and from crop (coming from IPK Gatersleben). Project description Today, the visualization and simulation of complex metabolic networks is of interest and importance for the future of Biotechnology (Systems Biology). The idea of this project is to bring together promising appendages coming from database integration, visualization and simulation. All partners have developed different tools. Prof. Hofestädt is an expert in integration and Petri net simulation of networks. Prof. Schreiber is an expert in the visualization of networks and Prof. Chen has a strong background in pathway prediction and alignment. By combining our tools and experience, we are able to implement a powerful tool for the analysis of metabolic pathways based on the Petri net analysis and simulation. This tool can also be the backbone of a virtual cell simulator. The realization of a virtual cell is the focus of various national and international projects. Until now, no qualified theoretical concept and tool could be presented. Run time: 2008 - 2010 Project Members: AG Bioinformatik: Prof. Ralf Hofestädt IPK Gatersleben, Plant Bioinformatics Group: Prof. Dr. F. Schreiber, Dr. U. Scholz Zhejiang University, College of Life Sciences: Prof. Dr. M. Chen, Prof. Dr. Wu |
| BMBF Project: "Prediction and reconstruction of metabolic networks based on textmining methods" |
|
The cooperation between Prof. Kolchanov and Prof. Hofestädt has become a long tradition. Since 1998 we have various different seminars, workshops and summer schools together. One highlight of this cooperation is the foundation of the German/Russian network mentioned above. Furthermore, since 2000 Prof. Hofestädt has been the co-Chairman of the BGRS conference, which is organized regularly in Novosibirsk (next conference will be June 2008). Reading the textbook of Prof. Ratner (Novosibrisk) Prof. Hofestädt began to model metabolic networks based on formal languages at the beginning of the 80’s (as a student). His Phd thesis and his Habilitation were based on the idea to use formal languages for the description of metabolic processes and networks. During the second half of the 90’s, when Prof. Hofestädt was Professor at the University of Magdeburg, he worked more closely with Prof. Kolchanov. Molecular databases and integration was becoming more and more the topic of our research. Both of us developed different database systems and started to develop integration tools for the automatic access and integration of molecular data. One result of the activities in Novosibirks is the ANDVisio tool, which we would like to further develop togehter in this project. Bioinformatics, Systems Biology and Biotechnology are highly supported by DFG, BMBF and other national and international Foundations (VW-Stiftung etc.). However, we can see the same situation in Russia. During the last three years Prof. Hofestädt and Prof. Kolchanov founded the German/Russian Network Computational Systems Biology. All these activities were supported by BMB, Russian Ministry of Science and different other Partners. The future of Biotechnology and Molecular Biology will be strongly affected by the development of Bioinformatics and Systems Biology. Today the implementation of tools for the user specific integration of databases, analysis tools and information systems is the most important topic of Bioinformatics and Systems Biology. Already in the world wide web we can see more than 1000 molecular information systems representing information about genes, proteins, enzymes, pathways, etc. Behind these database systems thousands of analysis tools are available in the internet. Based on these information systems and analysis tools the user is looking for integrative tools, which will help to realize the user specific application of selected databases and analysis tools. The goal of our project seeks to offer a system, which is able to predict and analyse complex metabolic networks based on database integration and textmining tools. Therefore, we will systematically extend the ANDVisio tool. One point is the user interface and the other is to use methods of deductive databases to support the automatic generation of new molecular knowledge. For the user interface we would like to implement a 3D visualisation tool for the analysis of predicted metabolic networks. For the generation of new knowledge we would like to implement a rule based systems for the automatic generation of metabolic networks. We plan to cooperate from the beginning with Prof. Thiele (Bruker). He is also member of the German/Russian Network of Computational Systems Biology and interested in the results of this project. Few weeks ago Prof. Kolchanov presented ANDVisio in Bremen. Furthermore, we discussed the ideas of this project together in Bielefeld. Run time: 2008 - 2009 Project Members: AG Bioinformatik: Prof. Ralf Hofestädt, Dipl.-Inform. David Braun, Dipl.-Inform. Thorben Wallmeyer, M.A. Björn Sommer |
| German/Russian Virtual Network of Bioinformatics - Computational Systems Biology |
| Flyer available in English or in German |
|
DAAD PPP German-Norwegian Collaborative Research Support Scheme Project: "Modelling and simulation of Quorum Sensing in 3-dimensional space in V. salmonicida" (2007-2008) |
|
The intention of this work is to develop a basic simulation environment for simulating different types of bacterial behaviour for a better
understanding of Quorum Sensing. Therefore a virtual world will be implemented with different configurable parameters. The bacteria will be
represented as so called agents with a simplified cell-cycle, the ability of movement as well as detection of other bacterial agents in the
immediate vicinity. Also a configurable list of given reactions will simulate a basic intracellular network for each agent. Partner
Prof. Dr. Nils Peder Willassen
University of Tromsø People: Prof. Dr. Ralf Hofestädt, Dr. Thoralf Töpel. B.Sc. Sebastian Janowski |
| EU FP6: CardioWorkBench: Drug Design for Cardiovascular Diseases - Integration of In Silico and In Vitro Analysis |
| BMBF Joint Research Project: CELLECT: Databases and data integration |
|
Der Schwerpunkt dieses Teilprojektes liegt in der projektübergreifenden Bereitstellung von Methoden aus dem Bereich Datenbanksysteme, dezentrale Datenhaltung und die Integration externer Daten-banken. Im Verbundprojekt werden Lösungen zur Datenvernetzung unter den Projektpartnern erarbei-tet, wodurch ein Kommunikations- bzw. Datennetzwerk aufgebaut wird, was die gezielte Integration von Daten und Analysemethoden im Projekt unterstützt. Die aus den MELK-basierten Proteom-Analysen und weiteren Laborexperimenten gewonnenen Daten werden in einer zentralen Datenbank gespeichert und gepflegt. Der automatische Zugang und die Analyse der Daten steht den Projektpart-nern zentral zur Verfügung. Neben dem im Projekt gewonnen topologischen Proteomdaten präsentiert sich das heutige Wissen über molekulare Mechanismen in einem ständig wachsenden Datenbestand. Bereits heute sind über 300 molekularbiologische öffentliche Datenbanken via Internet verfügbar. In diesem Zusammenhang stellte die projektspezifische Nutzung dieser externen Datenbanken eine Möglichkeit dar, dieses Wis-sen in das Verbundprojekt einfließen zu lassen. Somit können die spezifischen Datenverarbeitungs- und Analysemethoden der Teilprojekte in einem weitaus globalerem Datenraum angewandt werden, was letztlich das Informationspotential erhöht. Die Berücksichtigung dieser externen Datenquellen er-höht die Aussage der Experimente und Analysen indem Querverbindungen und Zusammenhänge zu bereits vorhandenen Wissen aufgedeckt werden können. So können z.B. durch den automatischen Zugriff auf externe Datenbanken entsprechende Veröffentlichungen zu bestimmten Proteinstrukturen und -mustern, Signalketten, Vorhersagen von Proteintargets für bestimmte Krankheiten etc. ermittelt werden. Die relevanten Daten können dann anhand spezifischer Anforderungen extrahiert und bereit-gestellt werden. Die Notwendigkeit einer solchen Methode zur standardisierten Datenakquisition rückt verstärkt in den Vordergrund der aktuellen Forschungen der Bioinformatik. Neben der überwindung der physischen Datenverteilung und der damit verbundenen Heterogenität, ist die Berücksichtigung von nutzerspezifischen Anforderungen an integrierte Datenbestände ein wichtiger Aspekt. Dazu ge-hört auch die universelle Nutzbarkeit eines solchen Systems, was wiederum die Verwendung von standardisierten Schnittstellen und Anfragemöglichkeiten verlangt. Diese beiden Aspekte werden im Kontext der Teilprojekte bearbeitet und in Werkzeugen resultieren, die eine zentrale Kommunikations-, Analyse- und Informationsverarbeitungsplattform für alle Projekt-partner zur Verfügung stellt. Partner:
People:
|
| DFG-Forschergruppe Informationsfusion: Werkzeug zur Integration von Methoden und Daten zur DNA-Sequenzanalyse -Workbench für die Informationsfusion |
| DFG-Schwerpunkt: Informatikmethoden zur Analyse und Interpretation großer genomischer Datenmengen: "Modellierung und Animation regulatorischer Genwirknetze (MARG)" |
|
Number of grant (DFG): HO 1178/10-1 Project Leader: Ralf Hofestädt, Faculty of Technology, Bielefeld University, D-33619 Bielefeld Scientists: Ming Chen, Andreas Freier, Matthias Lange, Jacob Köhler Background of project: Although substantial progress has been achieved within the application of database and internet technology in the field of molecular biology, the problem of integrating distributed data as a preprocessing step for the analysis of genomic datasetsis still a challenging task. Actually, there are hundreds of distributed and heterogeneous databases [Galperin], where genomic information is stored. However, existing file formats, interfaces and databases change over time, which makes it a permanent problem to access and integrate the recent datasources. To overcome this, universal and integrative tools are needed.
For the modelling and analysis of metabolic, protein and gene gene networks, access to different pathway, protein interaction, gene regulatory and other
databases is essential. Partially overlapping datasets have to be fused in order to integrate networks of different types. For this reason, it is a typical
data integration task. General references related to project:
Aims of project: The aim of the project was to develop a process to integratively design models of metabolic networks. Furthermore, a software platform needed to be implemented, providing generic tools to apply the developed methods to user-specific biological tasks. Our project places emphasis on two points:
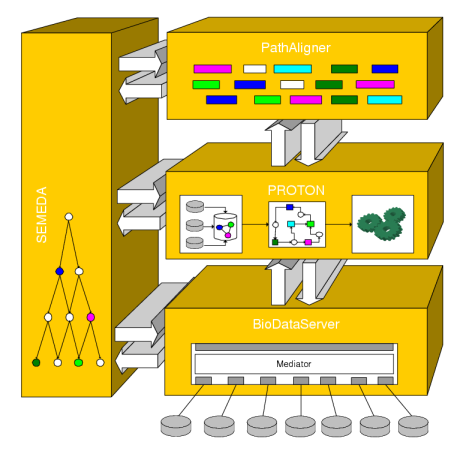
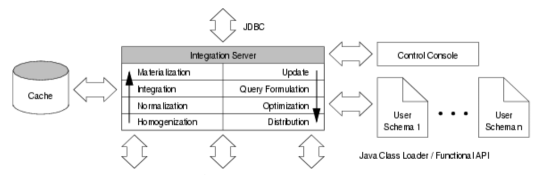
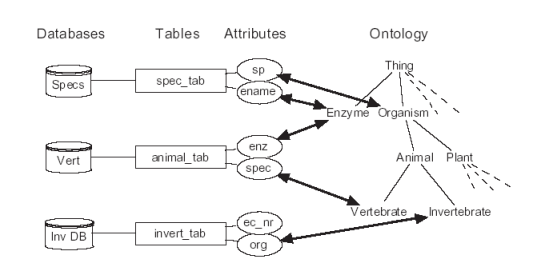
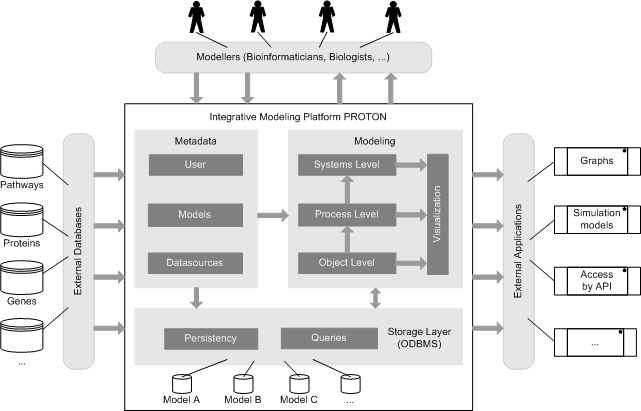
Description of project results: In the center of our approach we developed a process of what we call Integrative Modelling of Biochemical Networks. As an implementation of the process, we implemented the software plattform MARGBench, which is named after this project. Figure 1 shows the projects overall architecture.
Figure 1 MARGBench includes the following four modules:
Publications of project results:
Organized Workshops (DFG supported):
|
| BMBF Deutsches Humangenomprojekt (DHGP): "Integrative Simulation of Gene Controlled Biochemical Networks" |
|
Grants (BMBF): 01KW9912 (01.11.1999-31.10.2002), 01KW0202 (01.11.2002-31.08.2004) Project Leader: Prof. Dr. Ralf Hofestädt, Bielefeld University, Bioinformatics/Medical Informatics Department Scientists: Niels Grabe, Matthias Lange, Thoralf Töpel Background: Today we face a number of valuable data sources giving specialized views to highly specific aspects of biological systems ranging from genetic sequences up to pathological data. The same applies to the corresponding software where programs for analysis, modeling and simulation of individual aspects of biological data have been developed. These facts lead to the general task of integrating all this knowledge to make it biotechnologically and medically applicable. Aim: The goal of our project is to implement an information system which supports the analysis of gene controlled biochemical networks. Using and enhancing existing methods of bioinformatics, we plan to integrate different biochemical, molecular and medical databases which are important for the analysis of metabolic diseases. A special data integration system, partially based on the design of federated database systems and on the prototype of our Biobench system will be designed. As a second step methods for integration of tools, simulating gene controlled biochemical networks will enhance this system and lead to an integrated server for gene controlled biochemical networks. Method: Petrinet and rule-based simulation methods will be studied for modeling and simulation cellular signal pathways based on cell communication and gene regulation processes. Promoter detection methods will be used from the research partner T. Werner. Integration of databases will be done using FDBS techniques. While TRANSFAC contains knowledge about gene expression, biochemical reactions and signal pathways are supplied by KEGG and TRANSPATH. BRENDA stores behavior of enzymatic driven processes. For medical data METAGENE and MDDB are used. The simulation tools and databases will be combined into a gene regulation server accessible via web interface. Result: The developed Internet server establishes a widely usable system concerning gene regulation starting on the mere sequence level up to pathological data. The project reshapes the currently available biomedical knowledge by integrating it, eases its understanding and thus opens up the road for exploiting it for medical and pharmaceutical research. |
| Expertensystem D3-IEM - Unterstützung bei Untersuchungsanforderung, Diagnose und Therapie |
| Visualisierung und Animation Metabolischer Prozesse |
| HIEMIS - Human Inborn Errors of Metabolism Information System - Wissensrepräsentation im Internet |
| Kooperation mit dem Fraunhofer-Institut für Aerosol und Toxikologieforschung in Hannover |
Stifterverband für die Deutsche Wissenschaft (Kurt-Eberhard-Bode Stiftung):
|
| Landesprojekt des Landes Sachsen-Anhalt: Molekularer Wissensserver der Genregulation |
| VW-Stiftung: Modellierung und Simulation der Genregulation |
| Knowledge Discovery zur Sequenzanalyse - Ali Baba |
| Entwurf und Implementierung eines Informationssystems zur Verwaltung molekularbiologischer Simulationsdaten (MEKDB) |
| Entwicklung eines Wissensservers für die innere Medizin |
| BMBF-Förderinitiative Notebook University im Programm "Neue Medien in der Bildung" |